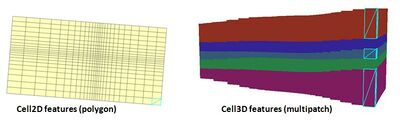
AHGW:Build Cell3D Features
The Build Cell3D tool is part of the MODFLOW analyst toolset. The tool creates 3D cells (multipatches) based on existing Cell2D features (polygons) and information on the discretization of the MODFLOW grid stored in the modflow data model tables.
Data from the Discretization and Basic components are used to create three-dimensional Cell3D features from two-dimensional Cell2D polygons.
The following tables from the Discretization (DIS) component are used by the Build Cell3D tool:
DISVars – Defines the model dimension. The following fields must be populated:
- NLAY: number of layers.
- NROW: number of rows.
- NCOL: number of columns.
TopElev – Contains the top elevations of the model cells:
- TopElev: top elevation value must be specified for each active cell in the top layer (layer 1). It is optional to have elevation values for inactive cells as well.
BotmElev – Contains the bottom elevations of cells and confining beds (CBD):
- BotmElev: bottom elevation must be defined for each active cell (optional to also have elevations for inactive cells).
- BotmElevCBD: represents the bottom elevation of confining beds (optional).
DISLayers – Contains layer multipliers. The table should be fully populated (a row for each layer in the model):
- LAYCBD: flag for turning on/off confining beds. LAYCBD = 0 confining beds are turned off, LAYCBD = 1 confining beds are turned on.
- AM_TopElev: top elevation array multipler.
- AM_BotmElev: bottom elevation array multiplier.
- AM_BotmElevCBD: bottom elevation of confining beds array multiplier.
The following tables from the Basic (BAS6) component are used by the Build Cell3D tool:
Basic – Contains the IBOUND and starting head arrays:
- IBOUND: defines if cells are active (IBOUND > 0), Inactive (IBOUND = 0), or constant head cells (IBOUND < 0). An IBOUND value should exist for each cell in the model.
BasicArrayMult – Contains IBOUND and starting head layer multipliers. For each layer of the model an IBOUND multiplier should be populated. This value can be used to activate/deactivate selected layers:
- IBOUND: A layer multiplier that is appliaed to IBOUND values from the Basic table.
Usage Tips
- Make sure the Cell2D and Cell3D feature classes have the same length units as the modflow model.
Command Line Syntax
BuildCell3D <in_cell2d> <in_cell3d> <in_gdb> {active_cells | all_cells}
Parameters
- Cell2D feature class
- Cell3D feature class
- Geodatabase file containing the modflow data model tables.
- Create Cell3D features for all cells or only active cells (default only active cells)
Examples
Command Line:
BuildCell3D Cell2D Cell3D in_gdb2 active_cells
Script:
# Build Cell3D features
# Requirements: None
# Date: 03/21/2008
# Import system modules
import arcgisscripting, sys, os
# Create the Geoprocessor object
gp = arcgisscripting.create()
gp.AddToolbox("C:/ArcHydroGroundwater/ArcHydroGroundwater.tbx ")
try:
# set the Cell2D feature class
In_cell2d = “C:/Aquaveo/MODFLOW_Simulation.mdb/Cell2D”
# set the Cell3D feature class
In_cell3d = "C:/Aquaveo/MODFLOW_Simulation.mdb/Cell3D"
# set the input geodatabase
in_gdb = "C:/Aquaveo/MODFLOW_Simulation.mdb"
# Set the Boundary feature class
in_active = “true”
gp.BuildCell3D(in_cell2d, in_cell3d, in_gdb, in_active)
except:
# If an error occurred while running the tool, then print the messages
print gp.GetMessages()
See also